Software
I develop software as part of my research and as a hobby. I love working on Open Source projects. I believe that science must be open to everyone!
Below you’ll find packages/software/scripts that I developed (some of them were made in collaboration).
pymfe: Python Meta-Feature Extractor
The pymfe (python meta-feature extractor) provides a comprehensive set of meta-features implemented in python. The package brings cutting edge meta-features, following recent literature propose. The pymfe architecture was thought to systematically make the extraction, which can produce a robust set of meta-features. Moreover, pymfe follows recent meta-feature formalization aiming to make MtL reproducible.
The installation process is similar to other packages available on pip:
pip install -U pymfe
Example of usage:
# Load a dataset
from sklearn.datasets import load_iris
from pymfe.mfe import MFE
data = load_iris()
y = data.target
X = data.data
# Extract default measures
mfe = MFE()
mfe.fit(X, y)
ft = mfe.extract()
print(ft)
# Extract general, statistical and information-theoretic measures
mfe = MFE(groups=["general", "statistical", "info-theory"])
mfe.fit(X, y)
ft = mfe.extract()
print(ft)
# Extract all available measures
mfe = MFE(groups="all")
mfe.fit(X, y)
ft = mfe.extract()
print(ft)
We write a great Documentation to guide you on how to use the pymfe library.
Source code: https://github.com/ealcobaca/pymfe
System for Recommending Machine Learning Algorithms for Gene Expression Data Analysis Using Meta-learning
Cancer is one of the main causes of death today. Understanding its internal mechanisms and designing computational models capable of improving its diagnosis will have substantial benefits. New sequencing technologies, based on RNA-Seq, have made available a large amount of data that can be used for cancer diagnosis. As the manual analysis of these data is unfeasible, machine learning algorithms have been used successfully. However, each machine learning algorithm has an inductive bias, making it better suited to a given subset of problems. This project studies the use of strategies that improve the selection of classification algorithms for machine learning in the context of data classification. We investigate the potential of using meta-learning to associate characteristics present in a dataset with the most appropriate classification techniques to deal with them in identifying tumors through gene expression, using RNA-Seq and Microarray technology.
Source code: https://github.com/ealcobaca/rec-sys-to-cancer
Machine Learning for Glass Science (MLGlass)
In this repository, we keep some of the scripts and data used during our investigation about glass property prediction and design of new glasses. We have developed machine learning models capable of predicting the most common properties of glass with high performance. In addition, we created several scripts to optimize the models generated by the machine learning algorithms and to understand the knowledge acquired by these models (explainability).
Source code: https://github.com/ealcobaca/mlglass
Alchemist
The Alchemist uses a pool of optimization algorithms and machine learning models to search for viable glass compositions for a given glass transition temperature (Tg) value.

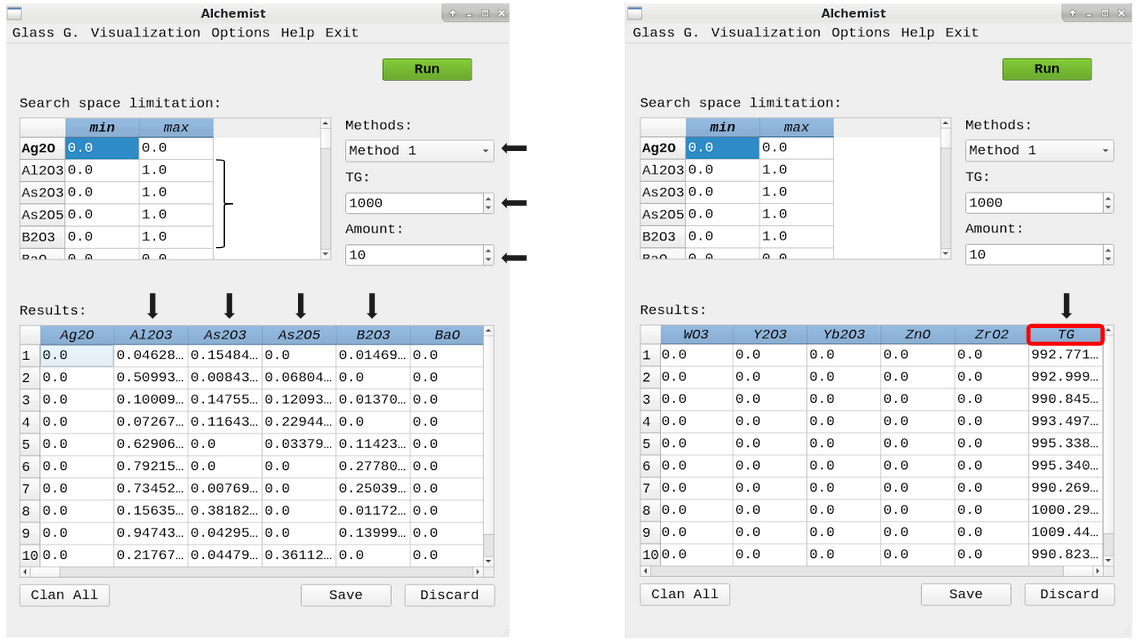
Source code: https://github.com/ealcobaca/alchemist-ia
Alchemist-Web
A Web based plataform for Alchemist.
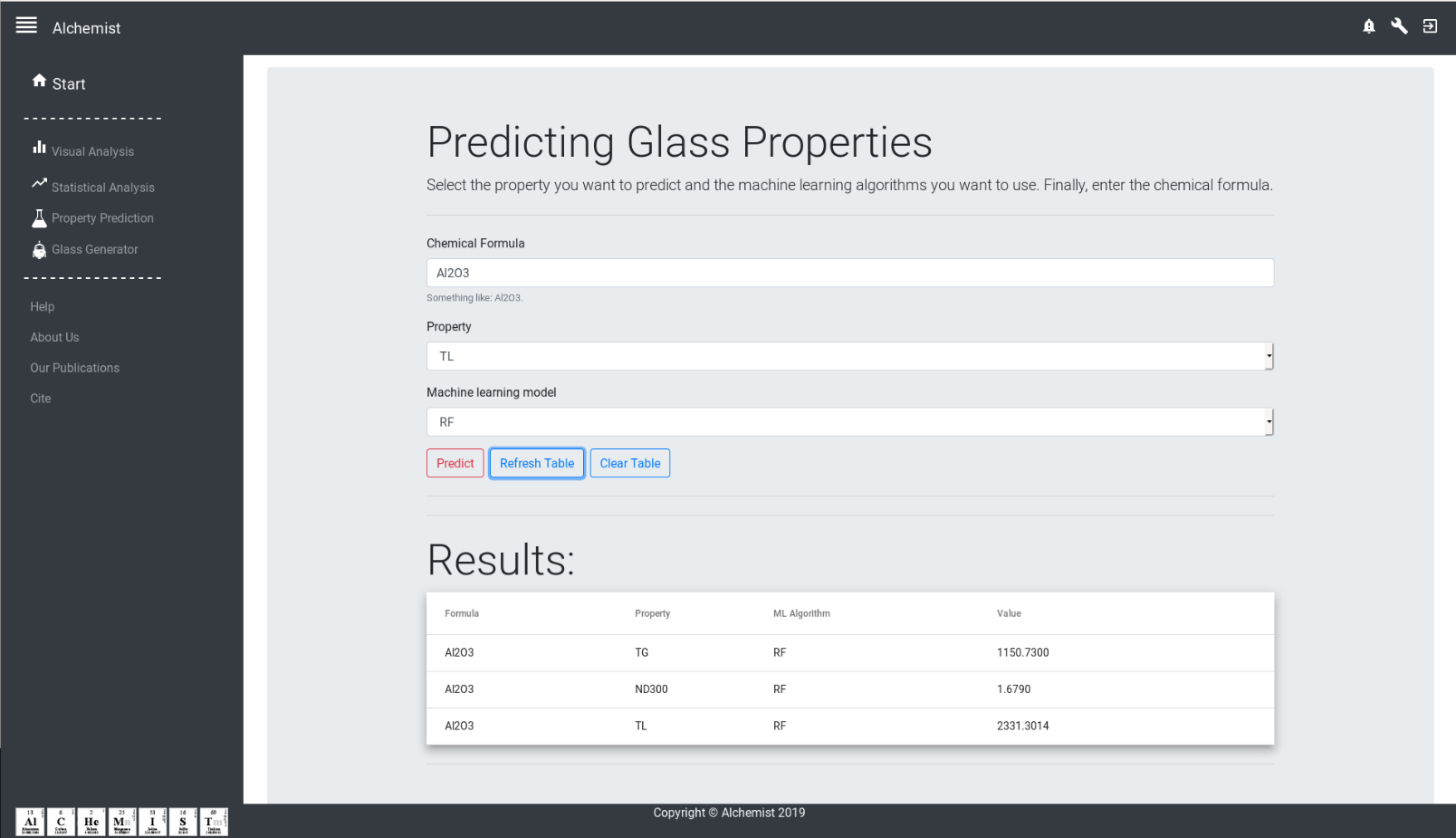
Source code: https://github.com/ealcobaca/alchemist-web
GeneVis
It is a framework for visualization gene expression data, gene/micro-RNA/RNA selection and tumor tissue classification using a perception-driven approach.
Source code: https://github.com/ealcobaca/GeneVis
Starving Wars
This video is a demo of the Starving Wars project carried out in the Computer Graphics course at ICMC - University of São Paulo.


